Diving into medical AI, I wanted to build something that could actually make a difference outside the lab. That's how ScopeAI was born: a real-time, lightweight polyp detection tool for colonoscopy images, designed specifically for clinics with limited compute. Here's the full story-from patch-based CNNs to YOLOv8, and all the lessons learned along the way.
Why Build ScopeAI?
Colorectal cancer is one of the most common cancers globally, and early detection of polyps during colonoscopy can save lives. But most AI solutions need powerful GPUs and aren't practical for rural clinics or small hospitals.
Goal: Build an accurate, interpretable polyp detector that runs in real time, even on a mid-range laptop or desktop.
Meet ScopeAI: The User Experience
Home Screen
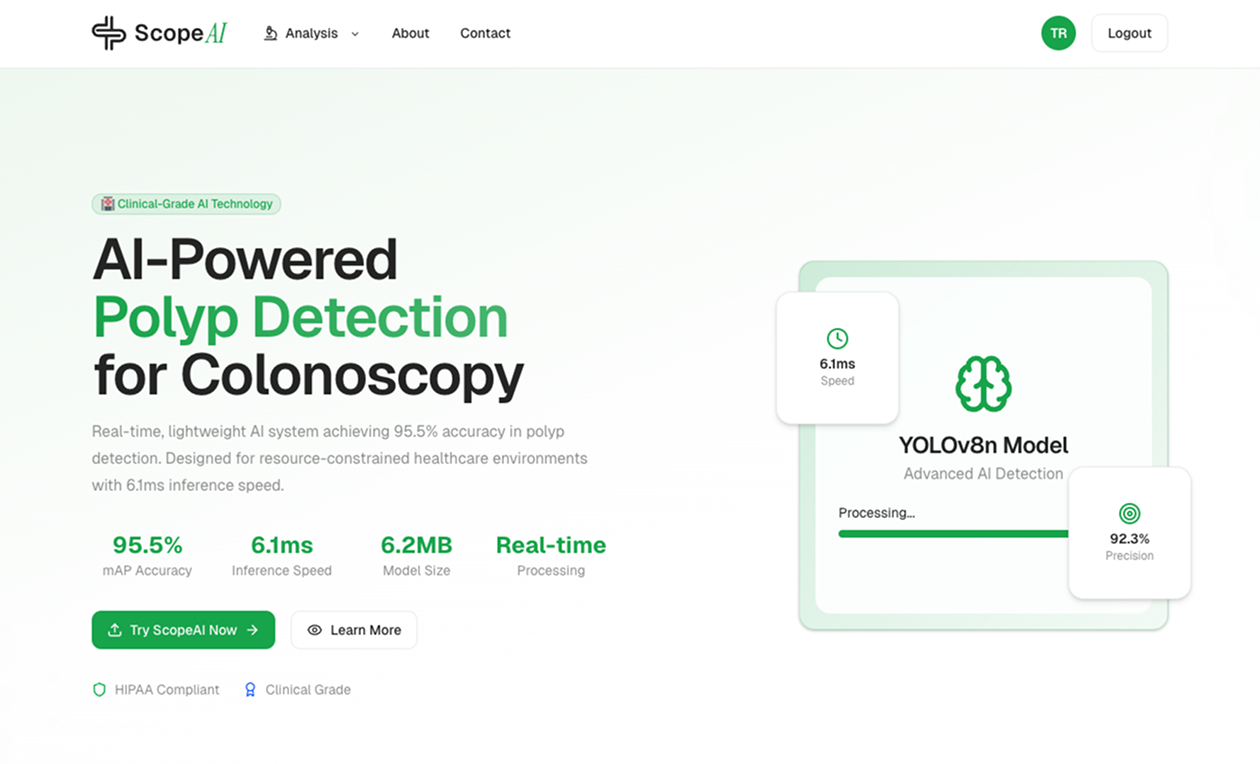 The ScopeAI landing page-simple, intuitive, and ready for analysis.
The ScopeAI landing page-simple, intuitive, and ready for analysis.
ScopeAI was designed to be approachable for clinicians and researchers alike. The home page welcomes users with clear navigation and a streamlined workflow.
Login & Register

User authentication keeps data secure and allows for personalized analysis history.
Profile & History
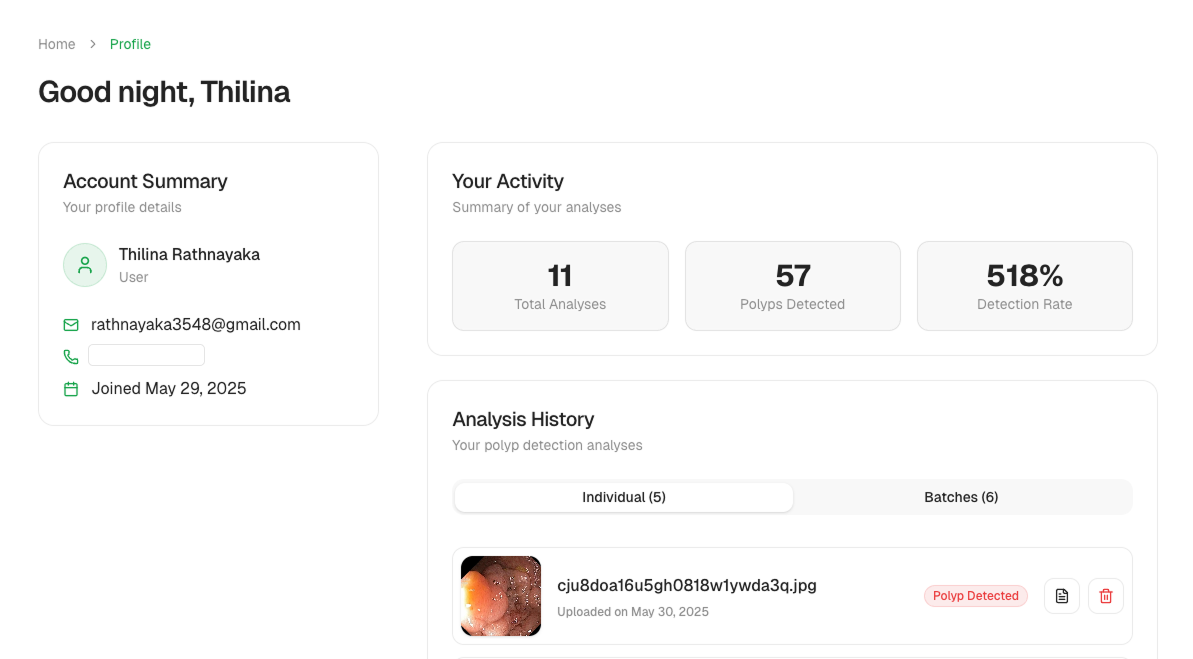
Each user can review past analyses, revisit reports, and manage their account.
Project Evolution: From Sliding Windows to YOLOv8
The Patch-Based CNN Era
My first approach was a classic: sliding a window across the image, classifying each patch with a tiny CNN, and picking the highest-confidence region.
for window in sliding_windows(image, size=150, stride=10):
score = cnn_model.predict(window)
if score > best_score:
best_window = window
best_score = scorePros:
- Runs on almost any hardware
- No need for bounding box labels, just patch-level classification
- Easy to debug, explain, and visualize
Cons:
- Slow (2-3 seconds per image, even with batching)
- Only finds one polyp at a time
- Fixed-size bounding boxes
Enter YOLOv8: Real-Time, Multi-Polyp Detection
After benchmarking, I realized that with careful tuning, YOLOv8n (Nano) could run blazingly fast and deliver clinical-grade accuracy, even on modest hardware.
Key Upgrade: Swapped sliding window CNN for YOLOv8n: - Direct bounding box prediction - Multiple polyps per image - 6ms inference time - 95.5 mAP50 on Kvasir-SEG!
Dataset & Preprocessing
Primary Dataset: Kvasir-SEG
- 1,000 colonoscopy images with segmentation masks
Patch Extraction (for baseline CNN):
- Positive patches: centered over polyp masks
- Negative patches: random background or Kvasir v2 normals
YOLOv8 Training:
- Converted masks to bounding boxes
- 800 images for training, 200 for validation
train: ./images/train/
val: ./images/val/
nc: 1
names: ['polyp']Technical Stack
- Frontend: Next.js (React SPA)
- Backend: Flask (Python API)
- Model Training: Standalone scripts (TensorFlow/Keras for CNN, Ultralytics YOLOv8 for object detection)
- Database: SQLite (for upload/prediction history)
- Deployment: Local dual-server (Next.js on 3000, Flask on 5328)
api/
├── auth.py
├── database.py
├── inference.py
├── index.py
├── yolo_inference.py
├── models/
│ └── yolov8_polyp_best.pt
└── uploads/
README.md
requirements.txt
run.sh
uploads.dbAnalyzing with ScopeAI
Single Image Analysis
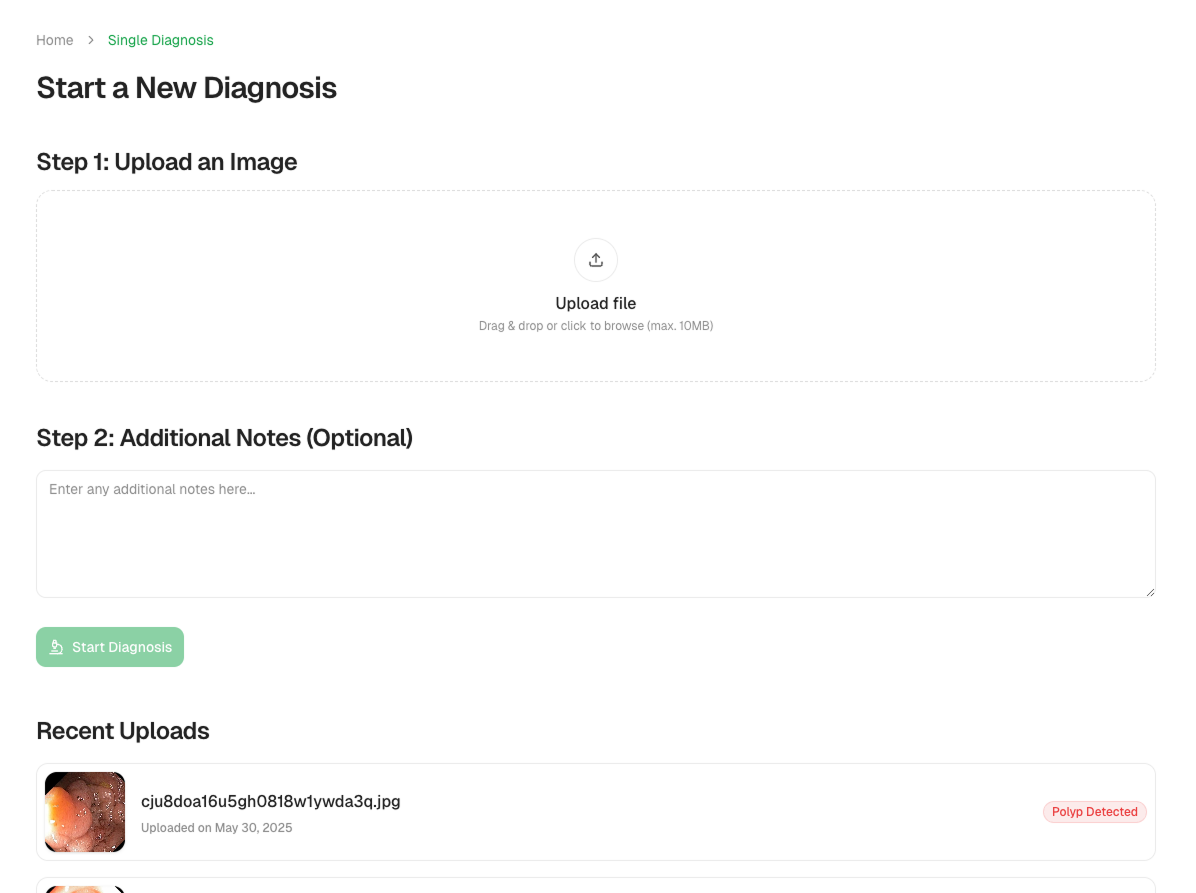
Just upload an image, and ScopeAI instantly detects and highlights polyps with bounding boxes and confidence scores.
Batch Analysis
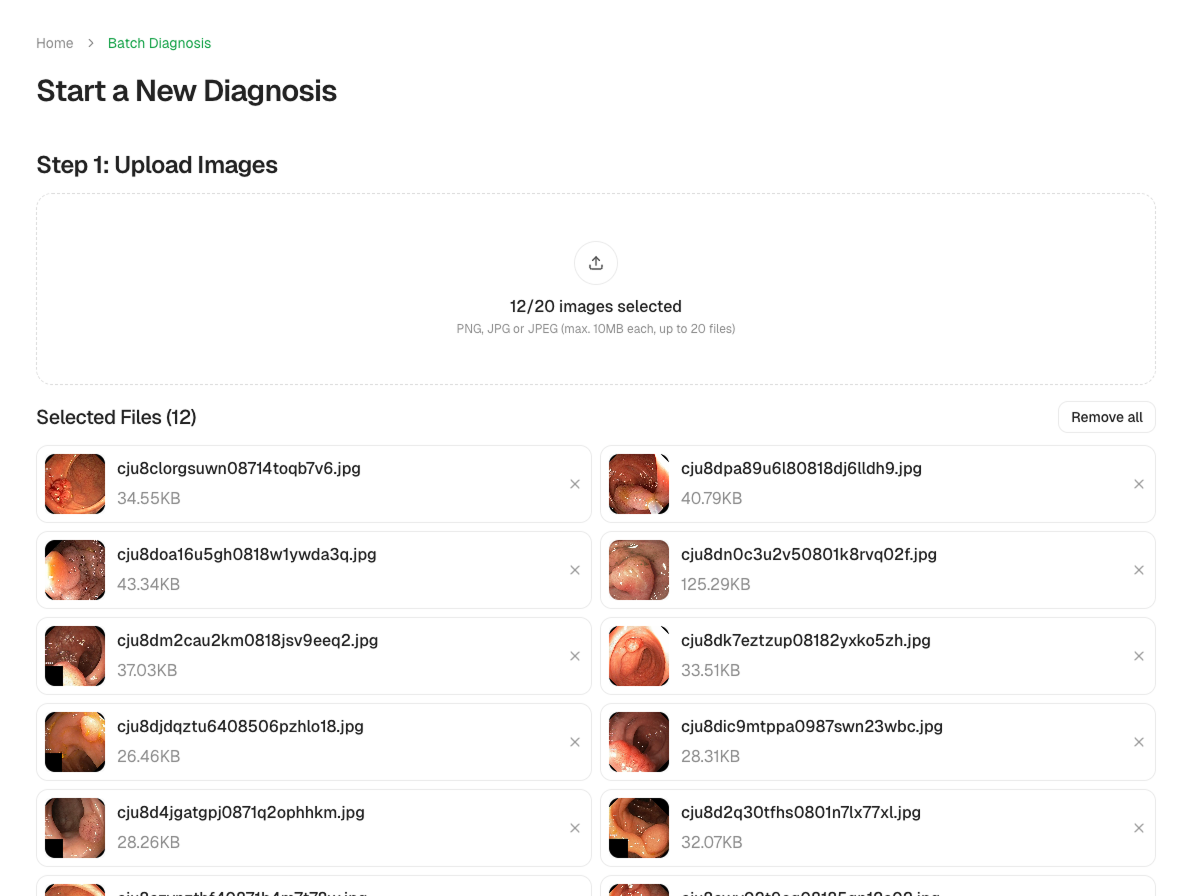
Process multiple images at once-ideal for research datasets or clinical workflows.
Analysis Report
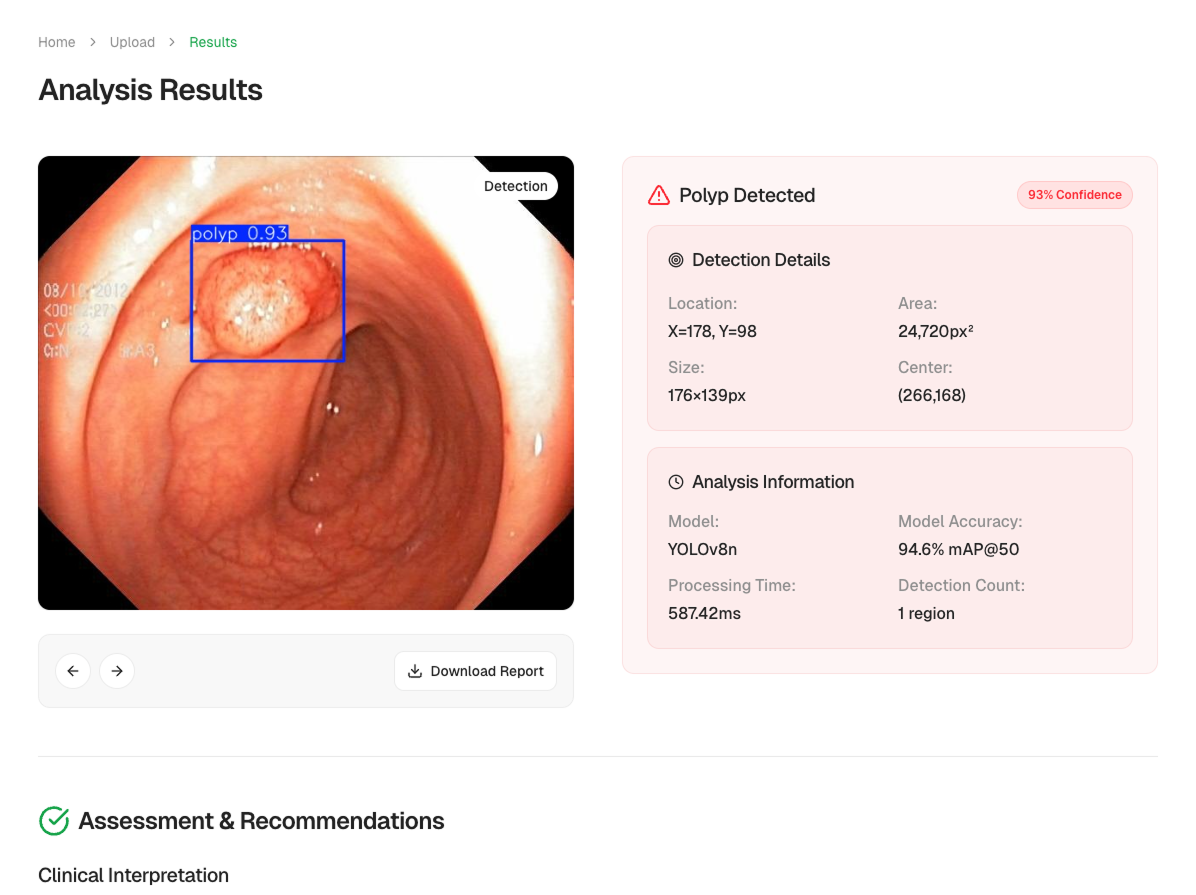
Each analysis comes with a downloadable report, including annotated images, detection statistics, and model confidence.
YOLOv8 Integration Details
Model Training
- Model: YOLOv8n (Nano, 3M params)
- Input: 640x640 RGB
- Epochs: 100 (best at epoch 100)
- Performance:
- mAP50: 95.5
- mAP50-95: 75.8
- Precision: 92.3
- Recall: 92.0
- Inference: ~6ms/image
- Model Size: 6.2MB
yolo train model=yolov8n.pt data=dataset.yaml epochs=100 imgsz=640 batch=16Flask API Integration
- Loads YOLOv8n model at startup
- Accepts image uploads, runs detection, and returns:
- Annotated image (bounding boxes)
- Heatmap overlay (confidence)
- Detection details (coordinates, confidence, count)
from ultralytics import YOLO
model = YOLO('models/yolov8_polyp_best.pt')
def yolodetection(image_path, confidence_threshold=0.5):
results = model.predict(image_path, conf=confidence_threshold) # process results, return annotated image, detections, heatmap, etc.Performance Comparison
| Approach | mAP50 | Inference Time | Multi-Polyp | Model Size |
|---|---|---|---|---|
| Sliding Window CNN | 88.2 | ~2.5s | No | 4.8MB |
| YOLOv8n (current) | 95.5 | ~6ms | Yes | 6.2MB |
Lessons Learned & Next Steps
- Sliding windows are great for prototyping, but YOLOv8 is a game-changer for real-world deployment.
- Careful dataset curation (especially negative samples) is crucial for robust detection.
- Even "Nano" models can hit clinical benchmarks when properly tuned.
What's next?
- Add video support (frame-by-frame detection)
- Grad-CAM overlays for explainability
- Pruning/quantization for mobile deployment
Project Availability
I'll be open-sourcing ScopeAI soon! Stay tuned for the code and documentation.
Who is ScopeAI For?
- Clinicians: Fast, interpretable detection for real-world screening.
- Researchers: Batch analysis, downloadable reports, and reproducible metrics.
- Students: Learn about practical deep learning deployment in healthcare.
Conclusion
Building ScopeAI taught me that with the right approach, you can bridge the gap between cutting-edge AI and practical, accessible healthcare tools. If you have questions, want to contribute, or just want to geek out about medical AI, drop a comment or reach out! Happy coding!